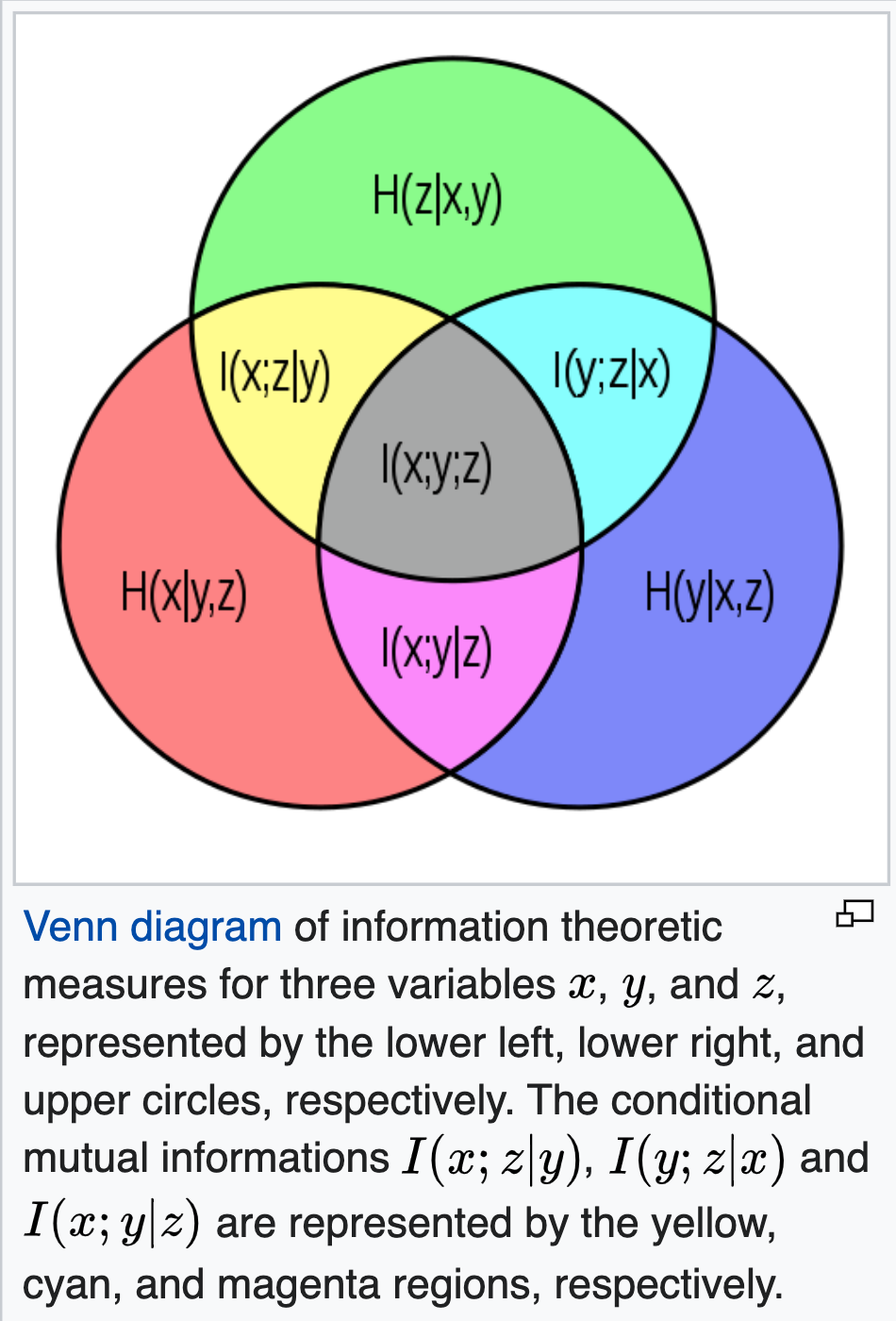
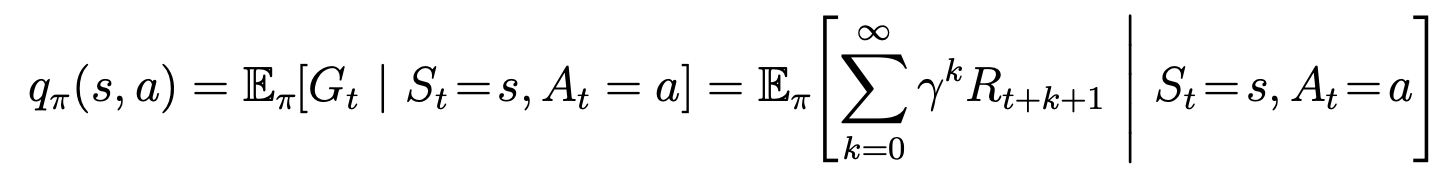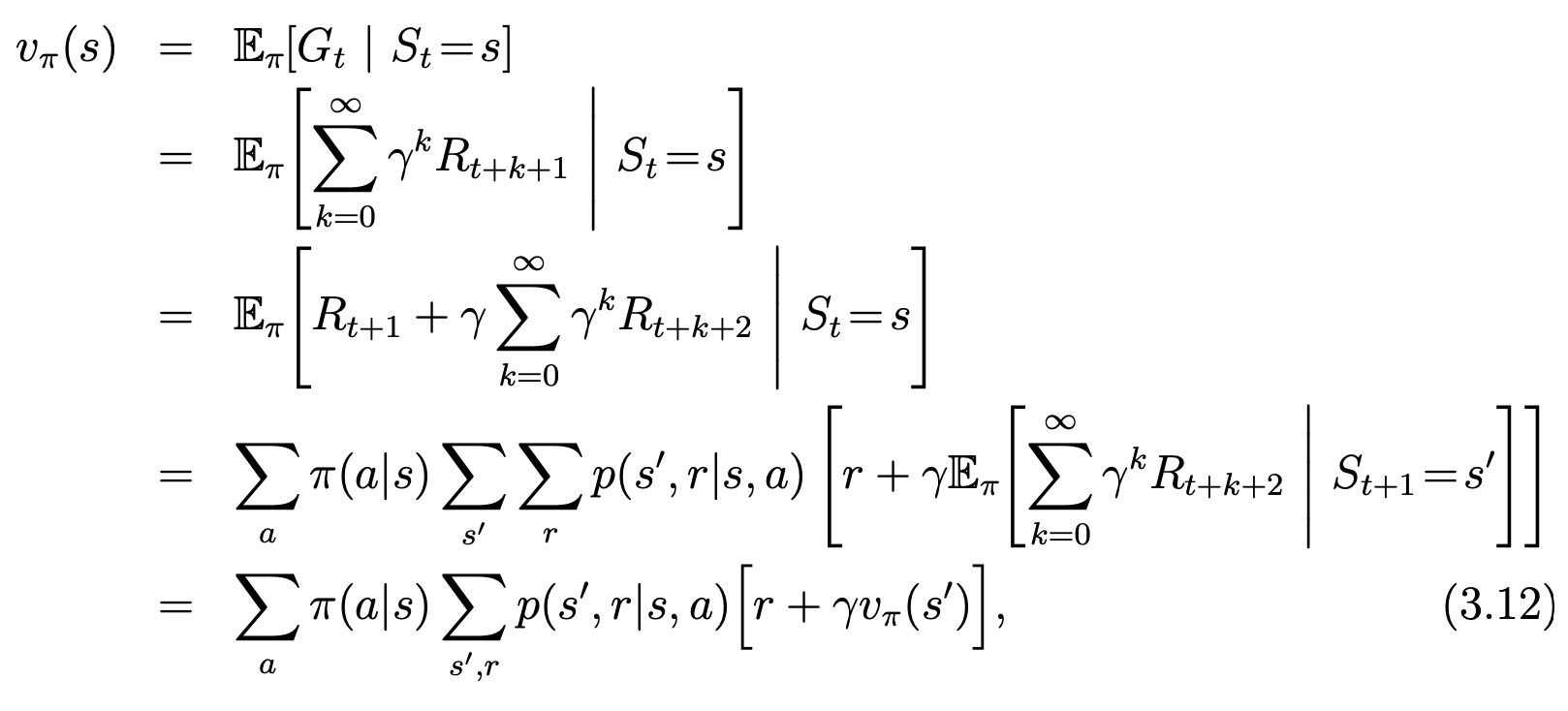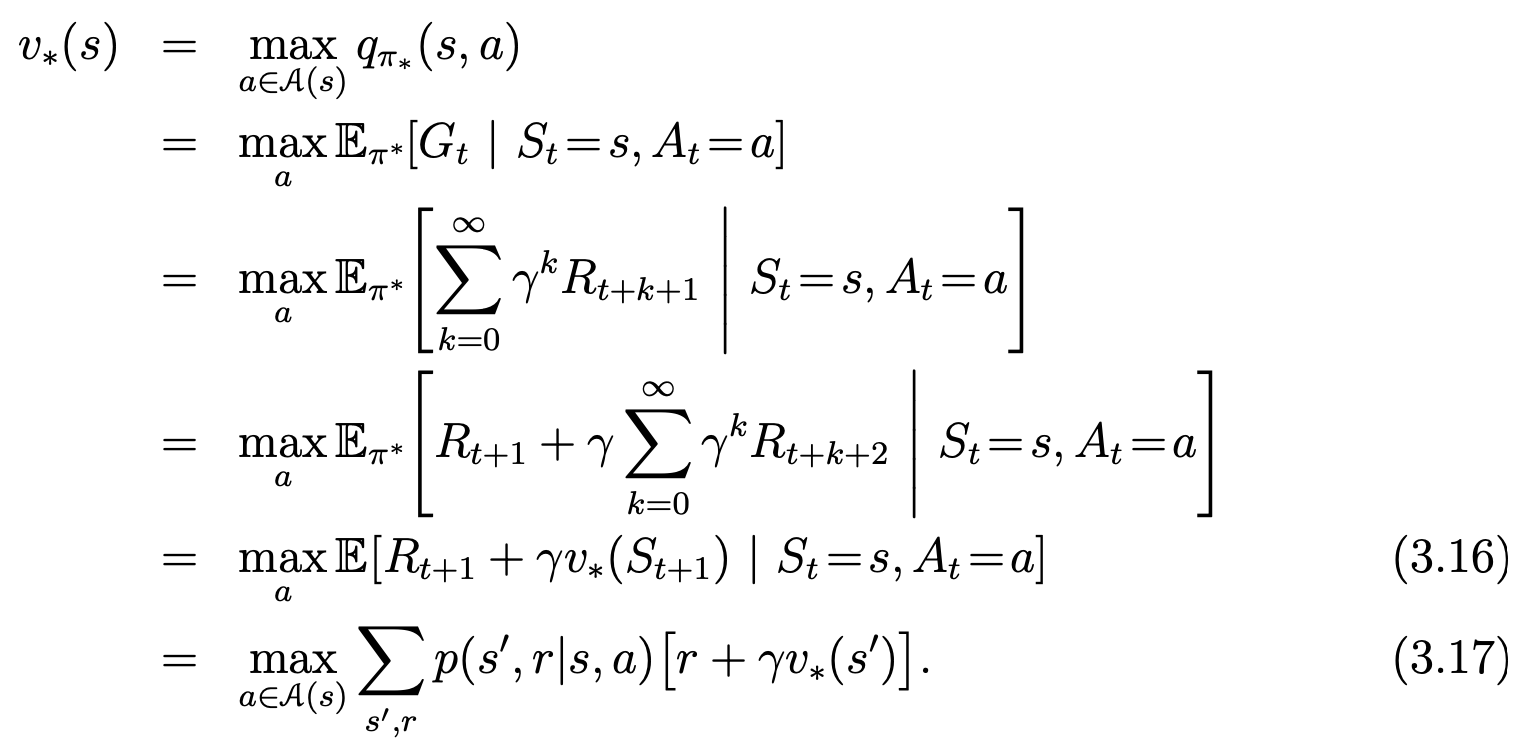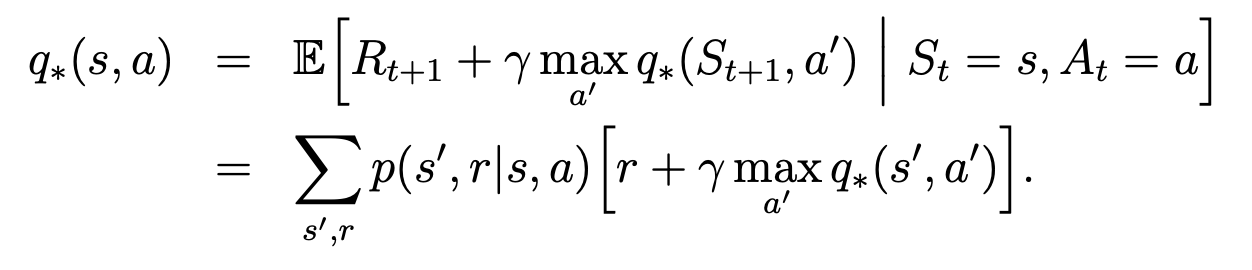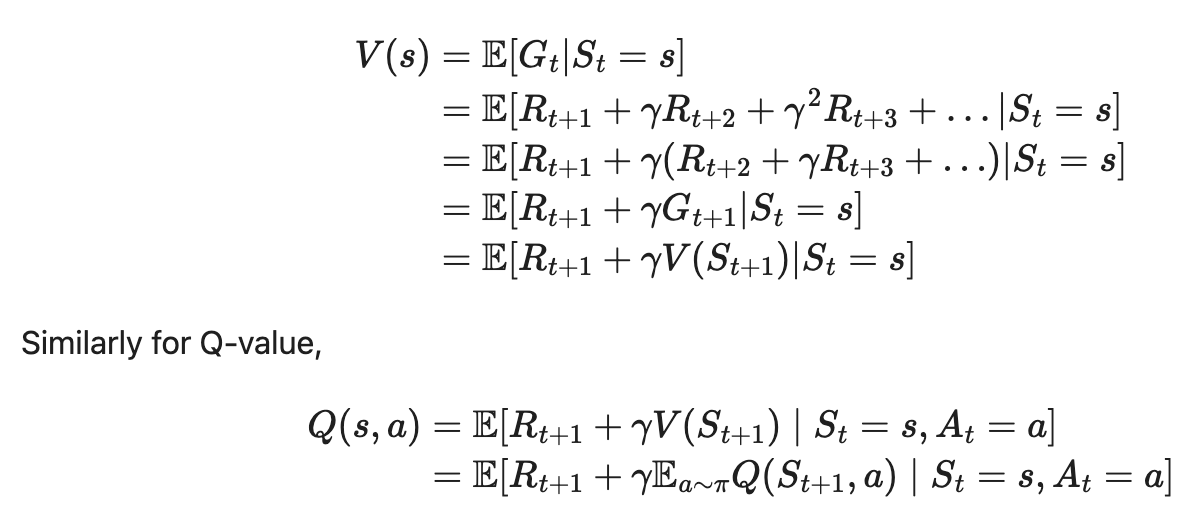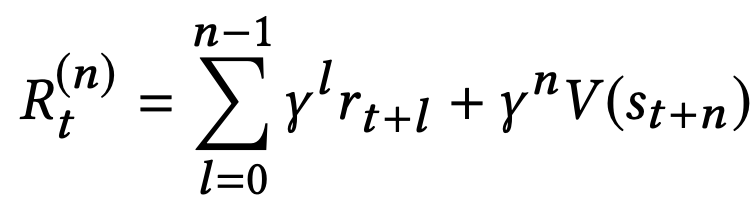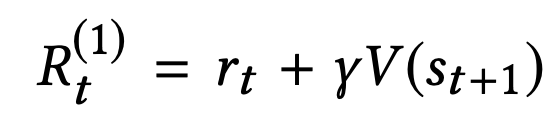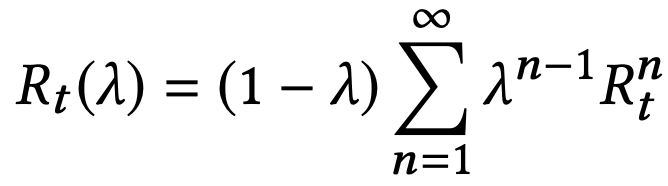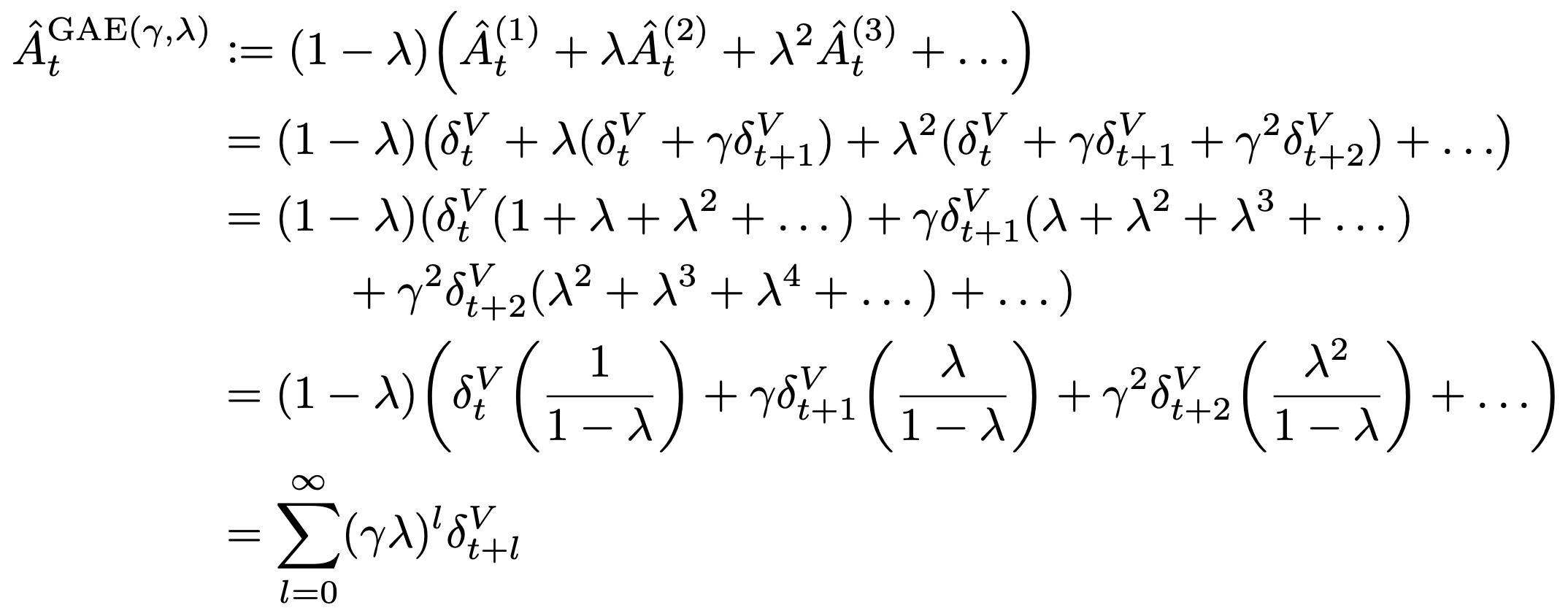中国大陆键盘布局(keyboard layout)和美国一样,即美式键盘(QWERTY)。
Chinese keyboard layout QWERTY [1]
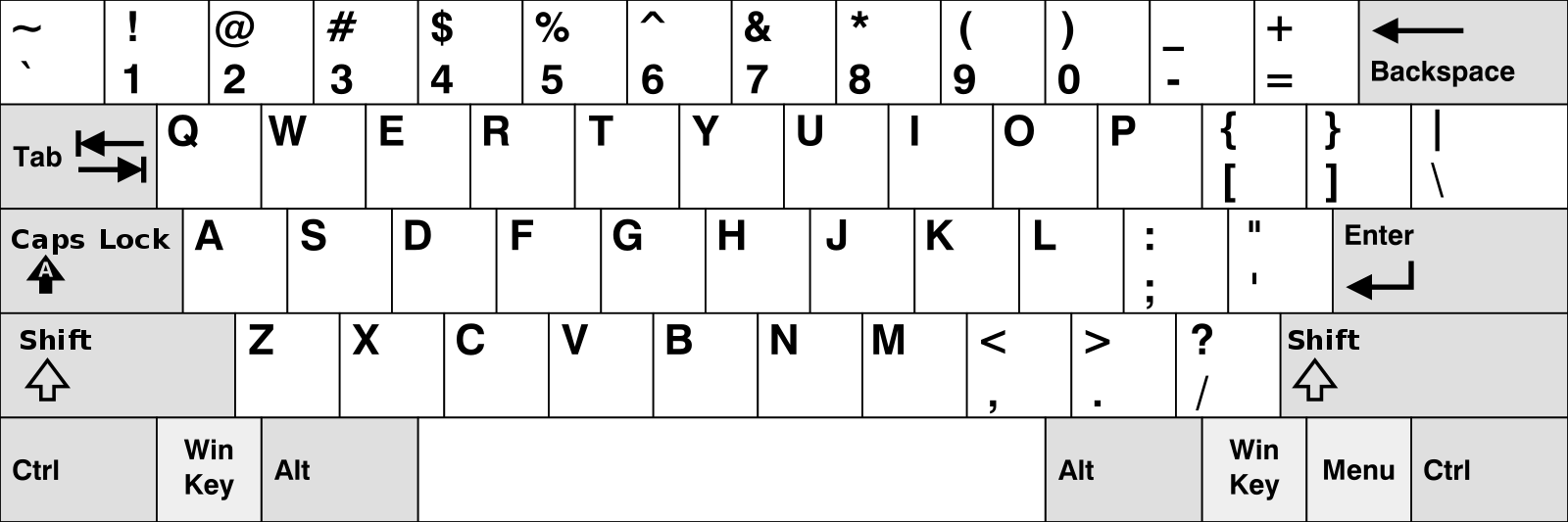
French keyboard layout AZERTY [1]

French keyboard layout AZERTY (MacBook or Apple keyboard) [2]

References
[1] 键盘布局. (2009, June 7). 维基百科,自由的百科全书. https://zh.wikipedia.org/wiki/%E9%94%AE%E7%9B%98%E5%B8%83%E5%B1%80
[2] Keyshorts. (2015, July 17). How to identify MacBook keyboard layout? https://keyshorts.com/blogs/blog/37615873-how-to-identify-macbook-keyboard-localization#french